Reprogramming Cancer Cells: A New Paradigm in Colon Cancer Treatment
- Cancer Reversion Therapy represents a dramatic shift in therapeutic philosophy.
- Colon cancer treatment has long focused on destroying malignant cells through surgery, chemotherapy, and other cytotoxic approaches. A groundbreaking new strategy instead aims to reprogram tumor cells back into normal cells, potentially overcoming drug resistance and side effects associated with cell-killing therapies.
- Recent research demonstrates that inhibiting a few key genetic “switches” can induce malignant colon cells to differentiate into benign intestinal cells, effectively reversing their cancerous state.
- If future clinical trials validate this strategy, it could inaugurate a new era of cancer treatment: one in which curing cancer does not always mean killing cells, but rather restoring the cellular order that cancer had disrupted.
Colon cancer is a prevalent malignancy and a leading cause of cancer-related deaths worldwide. Traditional colon cancer therapies share a common goal: eliminate the cancer cells, typically by surgical removal, toxic chemotherapy, or radiation.
Paradigm Shift: Cancer Reversion Therapy
One emerging concept is “cancer reversion” therapy – transforming cancer cells back into normal cells instead of destroying them. The idea is to re-educate malignant cells by flipping the internal genetic switches that determine cell identity and behavior. In late 2024, a research team at KAIST in South Korea provided a striking proof-of-concept: they developed a method to convert human colon cancer cells into normal-like colon cells without killing them. By targeting specific molecular regulators of cell fate, the scientists effectively reversed the malignant phenotype. The reprogrammed cells ceased uncontrolled growth and regained characteristics of healthy intestinal cells.
Achieving such cancer cell reversion required decoding the complex gene networks that govern how cells differentiate. The KAIST team built a high-resolution “digital twin” of the gene regulatory network underlying normal colon cell development. Using a computational framework called BENEIN (Boolean network inference and control), they analyzed thousands of single colon cells at various stages of maturation to map the switches that determine cell fate. In this model, each gene is treated as a binary switch (on/off) and the network of connections shows how genes collectively drive a cell toward a certain identity. By simulating different gene perturbations in silico, the researchers could predict which genes are master regulators of differentiation – the critical nodes that, when flipped, can redirect a cell’s trajectory.
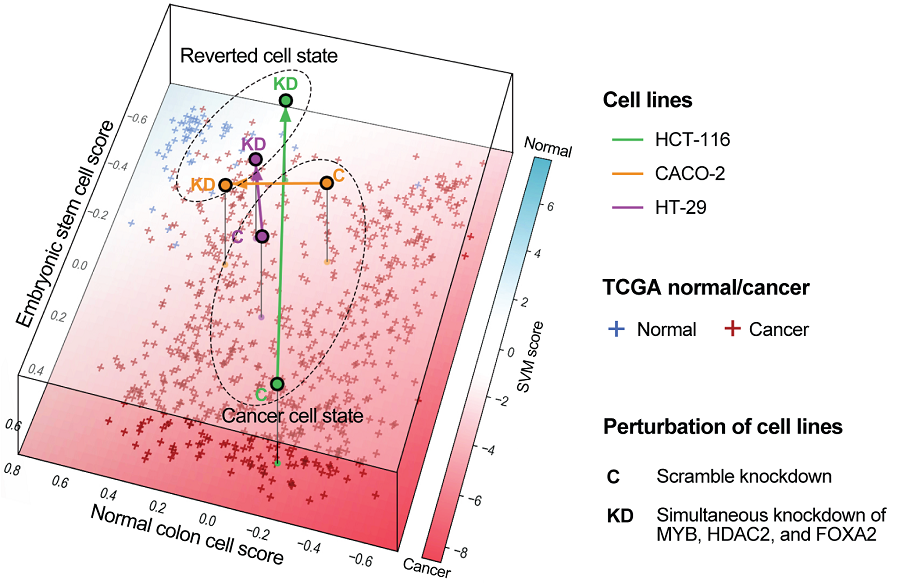
Through this systems biology approach, the team pinpointed a small set of genes that acted as top-level control switches. In fact, out of an initial network of 522 genes and ~1,841 interactions analyzed from 4,252 single cells, simulations revealed that just three genes held the reins on cell fate: MYB, HDAC2, and FOXA2. These genes were found to be preventing the cancer cells from maturing into specialized intestinal cells (enterocytes) and instead locking them in an undifferentiated, malignant state.

Each gene plays a distinct regulatory role:
- MYB is a transcription factor often overactive in colon tumors (and some leukemias) that promotes proliferation and blocks cellular maturation.
- HDAC2 is an epigenetic regulator (a histone deacetylase) that compacts DNA and silences tumor-suppressor genes, enabling cancer cells to sustain growth.
- FOXA2 is normally involved in developmental gene regulation, but in cancer it can be co-opted to support aberrant growth and survival signals.
By acting together, these three “master regulators” maintain the cancerous identity of colon cells..
Switching off cancer genes without DNA editing
With this knowledge, the researchers tested the effect of simultaneously switching off MYB, HDAC2, and FOXA2 in colorectal cancer cells. The result was striking: the cancer cells began to undergo differentiation. In laboratory cell cultures, blocking these three factors caused the malignant cells to slow their proliferation and lose invasive, stem-like traits. Morphologically and molecularly, they started to resemble normal enterocytes – the absorptive cells lining the colon.
Crucially, the reprogrammed cells showed a complete suppression of malignancy: they were no longer able to form aggressive tumors. In mouse models, human colon cancer cells treated to inactivate MYB/HDAC2/FOXA2 formed significantly smaller tumors than untreated cancer cells. This provided in vivo evidence that turning these switches “off” can tame the cancer. Furthermore, gene expression profiling confirmed that the reverted cells’ transcriptome closely matched that of normal healthy colon tissue. In essence, the cancer cells had been molecularly reset to a more benign, differentiated state.
Notably, this reprogramming was achieved without editing the cells’ DNA sequence. By using genetic and pharmacological inhibitors to dial down the activity of MYB, HDAC2, and FOXA2, the scientists avoided permanent genetic modifications. The cancer cells were guided back to normality through changes in gene expression alone leveraging the cell’s inherent plasticity.
Traditional treatments such as immunotherapy essentially wage war on tumor cells, an approach that can resemble a scorched-earth tactic. That is quite effective, but with considerable collateral damage. By contrast, the reprogramming strategy is more akin to negotiation or rehabilitation: it seeks to turn enemies into friends, converting dangerous cancer cells into normal cells that can safely coexist in the body. The potential advantages of this approach are profound. Because reversion therapy targets the regulatory causes of malignancy (the aberrant gene network states) rather than indiscriminately attacking rapidly dividing cells, it could spare healthy tissue and markedly reduce toxic side effects.
Another benefit is the possibility of preventing recurrence. Once a cancer cell has been truly normalized, it theoretically should lose its capacity to proliferate out of control or evolve into a resistant clone. In essence, there would be no “cancer” left to recur. This contrasts with standard treatments, which often leave behind a small reservoir of surviving tumor cells that can re-initiate growth. Early studies of the reprogramming method support this concept: treated cells showed durable changes, and tumors in mice were dramatically stunted without signs of new aggressive growth.
Universal solution for cancer reversion
The concept of a cancer reversion therapy – where cancer cells are not eradicated but rehabilitated – has profound clinical appeal. The proof-of-concept in colon cancer suggests that a similar strategy could be devised for other tumors by identifying their respective master regulators of differentiation.
In fact, the computational BENEIN framework is highly modular and data-driven, meaning it can be applied to single-cell gene expression data from different tissues to uncover key differentiation drivers in other cancer types.
As more single-cell atlases of human tissues become available, researchers could potentially map out “reversion roadmaps” for pancreatic, breast, lung cancers, and beyond. Each cancer might have its own set of switches whose inhibition prompts the tumor cells to revert to a more normal state.
From a commercialization standpoint, this breakthrough is already moving forward. The research team’s findings have been transferred to a startup to drive the development of practical cancer reversion therapies. The first applications are likely to be in colon cancer, aiming to complement or replace parts of standard treatment for patients with refractory disease.
Prepared by:

Jakub Knurek
Marketing Specialist
References
- KAIST Develops Foundational Technology to Revert Cancer Cells to Normal Cells, 2024.
- KAIST Discovers Molecular Switch that Reverses Cancerous Transformation at the Critical Moment of Transition, 2025.
- Oganezova T. South Korean Scientists at KAIST Reprogram Colon Cancer Cells in Breakthrough That Could Change Oncology. OncoDaily, 2025.
- Gong JR, Lee CK, Kim HM, Kim J, Jeon J, Park S, Cho KH. Control of Cellular Differentiation Trajectories for Cancer Reversion. Adv Sci (Weinh). 2025; 12(3): e2402132.
- Shin D, Gong JR, Jeong SD, Cho Y, Kim HP, Kim TY, Cho KH. Attractor Landscape Analysis Reveals a Reversion Switch in the Transition of Colorectal Tumorigenesis. Adv Sci (Weinh). 2025; 12(8): e2412503.
- Jung I, Hopper C, Jang SH, Yeo H, Cho KH. Reverse control of biological networks to restore phenotype landscapes. Sci Adv. 2025;11(34): eadw3995.
- Ku J, Jeong E, Gong JR, Cho KH, Sung CO, Kim SH. Identification of a unique subpopulation of mucosal fibroblasts in colorectal cancer with tumor-restraining characteristics. Mol Cells. 2025; 48(10): 100263.